Domestic dog origin of Carnivore Protoparvovirus 1 infection in a rescued free-ranging guiña (Leopardus guigna)
CONSERVATIONEMERGING DISEASESVIRUSWILDLIFE
Cite as: Rene Ortega, Juan Mena Vasquez, Sofía Grecco, et al. Domestic dog origin of Carnivore Protoparvovirus 1 infection in a rescued free-ranging guiña (Leopardus guigna). Authorea. May 22, 2020.
DOI: 10.22541/au.159018071.14311072
DOI: 10.22541/au.159018071.14311072
This is a preprint and has not been peer reviewed. Data may be preliminary.
Abstract
Carnivore protoparvovirus 1 is one of the most important pathogens affecting both wild and domestic carnivores. Here, we reported the genetic characterization of canine parvovirus strains from a rescued guiña (Leopardus guigna) and domestic dogs from Chile. Guiña sequence was classified as CPV-2c and phylogenetic analysis of the complete coding genome showed that the guiña CPV-2c strain share a recent common ancestor with Chilean domestic dogs strains. These viruses presented >99% identity and showed three changes in the NS1 protein, CHL-17 V596A, CHL-71 E661K and CHL-guigna L582F. This is the first detection and genetic characterization of CPV-2c infection in guiña worldwide and one of the few comparative studies that undoubtedly determine that the source of infection were domestic dogs. The current findings highlight that guiña is a susceptible species to protoparvovirus infection and that domestic dogs represent an important thread to its conservation. The CPV cross-species transmission between domestic dogs and guiña should be taken into account for protection programs of this endangerous species.
Title: Domestic dog origin of Carnivore Protoparvovirus 1 infection in a rescued free-ranging guiña (Leopardus guigna)
Running Head: CPV-2c infection in guiña
Authors: René Ortega1, Juan Mena4*, Sofía Grecco7, Rubén Pérez7, Yanina Panzera7, Constanza Napolitano5,6 Nhur-Aischa Zegpi1, Alberto Sandoval1, Daniel Sandoval1, Daniel González-Acuña2, Sergio Cofré3, Víctor Neira4 and Cristóbal Castillo-Aliaga1*.
*Co-corresponding authors
Affiliations:
1 Departamento de Patología y Medicina Preventiva, Facultad de Ciencias Veterinarias, Universidad de Concepción, Chillán, Chile.
2 Departamento de Ciencia Animal, Facultad de Ciencias Veterinarias, Universidad de Concepción, Chillán, Chile.
3 Departamento de Ciencias Clínicas, Facultad de Ciencias Veterinarias, Universidad de Concepción, Chillán, Chile.
4 Departamento de Medicina Preventiva Animal, Facultad de Ciencias Veterinarias y Pecuarias, Universidad de Chile, Santiago, Chile.
5 Departamento de Ciencias Biológicas y Biodiversidad, Universidad de Los Lagos, Osorno, Chile.
6. Instituto de Ecología y Biodiversidad (IEB), Santiago, Chile.
7 Sección Genética Evolutiva, Departamento de Biología Animal, Instituto de Biología, Facultad de Ciencias, Universidad de la República, Uruguay.
Summary
Carnivore protoparvovirus 1 is one of the most important pathogens affecting both wild and domestic carnivores. Here, we reported the genetic characterization of canine parvovirus strains from a rescued guiña (Leopardus guigna ) and domestic dogs from Chile. Guiña sequence was classified as CPV-2c and phylogenetic analysis of the complete coding genome showed that the guiña CPV-2c strain share a recent common ancestor with Chilean domestic dogs strains. These viruses presented >99% identity and showed three changes in the NS1 protein, CHL-17 V596A, CHL-71 E661K and CHL-guigna L582F. This is the first detection and genetic characterization of CPV-2c infection in guiña worldwide and one of the few comparative studies that undoubtedly determine that the source of infection were domestic dogs. The current findings highlight that guiña is a susceptible species to protoparvovirus infection and that domestic dogs represent an important thread to its conservation. The CPV cross-species transmission between domestic dogs and guiña should be taken into account for protection programs of this endangerous species.
Keywords: Canine parvovirus, Leopardus guigna , protoparvovirus 1, phylogeny, genetic characterization
Main Text
Introduction
Carnivore protoparvovirus 1 (CPPV-1) is a member of the genusProtoparvovirus of the family Parvoviridae . Like other parvoviruses, these are non-enveloped, small icosahedral viruses having a linear, single-stranded DNA genome of approximately 5.2 kb containing two major open reading frames (ORFs) (Cotmore et al., 2014). The first ORF encodes two non-structural proteins (NS1 and NS2), while the second ORF encodes the structural proteins VP1 and VP2 (Reed et al., 1988). CPPV-1 comprises several closely-related lineages previously considered different species, including feline parvovirus (FPV), canine parvovirus (CPV), mink enteritis virus, and raccoon parvovirus (Cotmore et al., 2014). These viruses are significant pathogens of veterinary relevance and affect both wild and domestic animals in the order Carnivore (Behdenna et al ., 2019; Chen et al ., 2019; Cotmore et al ., 2014; Cotmore et al ., 2019). A fatal CPV-2 infection was recently described in a rescued Taiwanese pangolin, providing the first evidence of CPV-2 infection in a non-carnivore species (Wang et al ., 2019).
FPV and CPV share a common recent ancestor that likely infected felids or a related wild carnivore species (Truyen et al ., 1992). When CPV-2 emerged, it only infected canids. This first CPV-2 type was soon replaced by a new lineage that regained the ability to infect felids and spread worldwide in few years. The high mutation capacity of CPV promoted the emergence of new genetic variants, including the antigenic/genetic variants known as 2a, 2b y 2c (Shackelton et al ., 2005). These variants recovered the ability to infect felines and started to pose a thread for non-dog species (Truyen et al ., 1996; Ikeda et al ., 2000).
In South-America, genetic and antigenic variants were described. The analysis of complete genome from several countries evidenced two migration events from Europe, an introduction from Asia and a lineage that likely diverged in South-America (Grecco et al ., 2018). Chile, Uruguay and Argentina have a similar scenario with the predominance of 2c strains of European origin (Castillo et al ., 2020) and the existence of 2a strains belonging to two different lineages (Pérez et al ., 2014; Gallo-Calderón et al ., 2015).
In wild species from South-America, there is antigenic evidence of the circulation of CPPV-1 in wild animals, including culpeo, grey and crab-eating foxes (Lycalopex culpaeus ; Lycalopex griseus; Cerdocyon thous ) (Martino et al ., 2004; Acosta-Jammet et al ., 2014), maned wolves (Chrysocyon brachyurus ) (de Almeida Curi et al ., 2012) and Geoffrey’s cats (Leopardus geoffroyi ) (Uhart et al ., 2012). Recently, a CPV-2c strain was isolated and genetically characterized from a dead coati (Nasua nasua) in Argentina (Bucafusco et al ., 2019).
Chile has a little information on the circulation of CPV in both domestic dogs and wild animals. Two serological studies evidenced the presence of antibodies against CPV-2 in domestic dogs and wild canids (Acosta-Jammet et al ., 2014; Acosta-Jammet et al ., 2015). A more recent study characterized the strains using VP2 sequence analysis and evidenced the existence of a predominant 2c variant of European origin (Castillo et al ., 2020).
The guiña (Leopardus guigna ) is a small felid inhabiting central and southern Chile and some areas in southwestern Argentina (Napolitanoet al ., 2014). According to the red list of the International Union for Conservation of Nature (IUCN), its conservation status is vulnerable, and the main causes are the reduction and fragmentation of its habitat (UICN, 2015). In recent years, feline viral immunodeficiency (FIV) and feline viral leukaemia (FeLV) infections have been reported in guiña populations and in both cases, phylogenetic analyses suggested a high association between wild and domestic animals viruses (Moraet al ., 2015).
The main objective of this study was to characterize the CPV in guiña and to establish the phylogenetic relationship with CPV strains obtained from domestic dogs inhabiting the near geographic area.
Materials and Methods
In July of 2015, a six-month-old sick guiña was recovered in an urban area from Curicó (Chile) by the Agricultural and Livestock Service (SAG) of Chile. The animal was submitted to the Center for Rehabilitation of Wild Fauna of the Universidad de Concepción (Chillán, Chile). The individual presented an intermittent hemorrhagic diarrhea, depression and anorexia during the clinical exam. A blood sample was collected to identify a possible infectious cause. Diagnostic test against FelV, FIV by Nested PCR (Mora et al., 2015) and against CPV by PCR were performed, resulting positive for CPV and negative to FeLV and FIV. A few weeks later, the animal died, and necropsy was carried out. Stool, stomach, small intestine and spleen tissues were collected.
DNA was extracted from blood sample using ThermoFisher Scientific DNA® Extraction from Blood (processed at the Virology Laboratory, Faculty of Veterinary Sciences, University of Concepción (Chillán, Chile)). DNA was extracted from tissue samples using DNeasy Blood and Tissue Kit and the QIAamp DNA Stool Mini Kit; Qiagen (Processed at the Molecular Biology Laboratory, Faculty of Sciences, University of Chile (Santiago, Chile)). The extracted genomic DNAs were stored at -20°C.
For CPV diagnosis, a small viral fragment of VP2 (583 bp) was amplified following Buonavoglia et al ., (2001). Furthermore, seven samples from Chilean domestic dogs were selected for complete coding genome sequencing. These samples were previously characterized by Castilloet al ., (2020) using sequencing of the VP2 gene. Therefore, the complete coding genome (4269 pb) from dogs and guiña were amplified following Pérez et al ., (2014).
The dataset consisted of sequences generated in this study and CPV complete coding genome sequences retrieved from the GenBank database. The filter criterion was to select the sequences according to its subtype, country and year of collection and host species other than dog. The sequences were aligned using MUSCLE v3.8.3 (Edgar, 2004) incorporated in AliView v1.26 (Larsson, 2014). The best-fit evolutionary model of nucleotide substitution was selected according to the Akaike Information Criterion (AIC) in jModelTest v2.1.6 software (Posada, 2008). Phylogenetic tree was inferred using the maximum likelihood method with RaxML-HPC2 v8.2.12 (Stamatakis, 2014) available in CIPRES Science Gateway (Miller et al ., 2010). The statistical support of the nodes was evaluated using 1000 bootstrap replicates.
Results and discussion
The blood sample of the guiña was CPV positive by PCR at the arrival to the Center for Rehabilitation of Wild Fauna. Post-mortem stool, stomach and spleen samples were also positive, confirming the CPV diagnosis and suggesting that the virus infected and established viremia in the host efficiently. The haemorrhagic diarrhoea and characteristic of the infected tissues collected in this case were similar to those found in protoparvovirus infections in other species (Allison et al ., 2013; Allison et al ., 2014; Oosthuizen et al ., 2019; Bucafusco et al ., 2019; Wang et al ., 2019; Viscardiet al ., 2019).
We obtained eight complete sequences of the CPV coding regions, one from the guiña and seven from domestic dogs. Details of these sequences are shown in Table 1. The guiña and Chilean dog sequences showed high genetic identity (99.8-99.9%). The sequence analysis of the VP2 region of the guiña sequence showed amino-acid changes associated with other 2c strains (Buonavoglia et al ., 2001; Decaro et al ., 2012), including 300G and 426E (Table 2). Three single nucleotide polymorphism lead to changes in the NS1 protein of three samples, CHL-17 V596A, CHL-71 E661K and CHL-guigna L582F (Table 3). Changes in the 582 NS1 position has been associated with neurological tissue tropism in FPV strains but the residue associated was different (L582S) (Gariglianyet al , 2016).
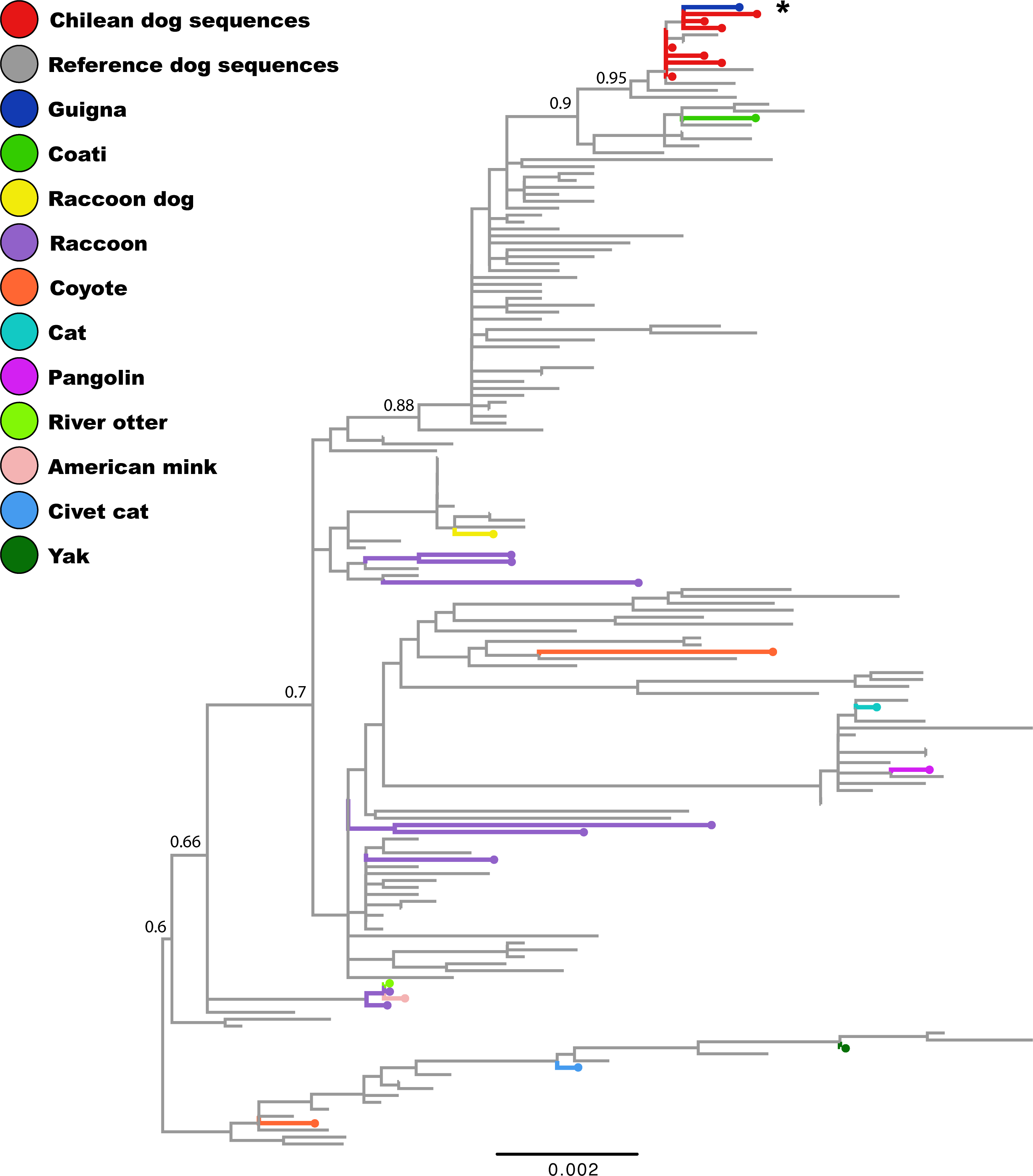
In South-America, Chile and Argentina have a predominance of 2c variant (Castillo et al ., 2020), but in Uruguay this variant is being replaced by a phylogenetically unrelated CPV-2a variant (Pérez et al ., 2014; Gallo-Calderón et al ., 2015). Phylogenetic analysis showed that the guiña parvovirus clustered with CPV-2c dog strains collected between 2009-2017 from Chile, Uruguay, Argentina, and Paraguay (Figure 1). Strains from guiña and coati diverged in different sub-clades, but both clustered in the Europe-I clade according to the recent classification by Grecco et al . (2018). Our results, suggests that the guiña virus is a recent acquisition of domestic dogs. Furthermore, apparently the 2c lineage may have a greater affinity for wildlife in South-America (Bucafusco et al ., 2019).
Several CPV infections in wild felids have been reported in Felis bengalensis (Nakamura et al ., 2001), Acinonyx jubatus ,Panthera tigris (Steinel et al ., 2000), Puma concolor , Lynx rufus (Allison et al ., 2013; Allisonet al ., 2014) and Leptailurus serval (Oosthuizen et al ., 2019). All of them showed high identity with domestic dog strains, and similar situation was described with the coati strain (Bucafuscoet al ., 2019). Recently, tiger parvovirus (TPV) has been described in captive tigers in China, and this has been the only variant described from a wild felid, showing greater similarity to FPV rather than to CPV (Wang et al ., 2019).
There are not many studies that show sequences comparison and analysis between domestic dogs and wild species (Chen et al., 2019; Oosthuizen et al ., 2019). Most reports only compare with sequences available in GenBank (Steinel et al ., 2000; Nakamuraet al., 2001; Allison et al., 2013; Allison et al ., 2014). For the conservation of this species, it will be important to consider the transmission of infectious agents from domestic animals. Likewise, infection by FeLV and FIV has been previously described in guiñas (Mora et al ., 2014). So, our finding might suggest that these animals would be at permanent risk of infection by these pathogens.
Acknowledgments
We are grateful to the staff of FISENLAB for all their support in the initial processing sampling, especially to Pedro Rojas. We thank the Molecular Diagnostic Laboratory of Department of Animal Pathology, especially to Álvaro Ruiz, for all their support in providing their facilities. We thank Dr. Francisco Espina Lopez for providing clinical cases.
Funding
This study was partly funded by project VRID Inicio, UdeC number 217.152.024-1.0 to RO, the Programa Fondecyt de Iniciación No. 11170877 to VN
Conflict of interest statement
The authors declare no conflict of interest
Data Availability Statement
The data that support the findings of this study are openly available in GenBank at Accession numbers MT458223, MT458224, MT458225, MT458226, MT458227, MT458228, MT458229 and MT458230.
Ethics Statement
The authors confirm that the ethical policies of the journal, as noted on the journal’s author guidelines page, have been adhered to and the appropriate ethical review committee approval has been received.
Was reviewed and approved by Comité de Bioetica de la Facultad de Ciencias Veterinarias, Universidad de Concepción. Dog samples was written informed consent for participation was not obtained from the owners because the animals were not identified individually. The guiña sample was taken during a clinical procedure and tissue samples were taken in the necropsy after the death.
References
- Acosta-Jamett, G., Cunningham, A. A., Bronsvoort, B. D., & Cleaveland, S. (2014). Serosurvey of canine distemper virus and canine parvovirus in wild canids and domestic dogs at the rural interface in the Coquimbo Region, Chile. European Journal of Wildlife Research, 61(2), 329-332.
- Acosta-Jamett, G., Surot, D., Cortés, M., Marambio, V., Valenzuela, C., Vallverdu, A., & Ward, M. P. (2015). Epidemiology of canine distemper and canine parvovirus in domestic dogs in urban and rural areas of the Araucanía region in Chile. Veterinary microbiology, 178(3-4), 260-264.2.
- Allison A. B., Kohler D. J., Fox K. A., Brown, J. D., Gerhold, R. W., Shearn-Bochsler, … Holmes, E. C. (2013) Frequent cross-species transmission of parvoviruses among diverse carnivore hosts. Journal of Virology, 87, 2342- 2347. https://doi.org/10.1128/JVI.02428-12
- Allison, A. B., Kohler, D. J., Ortega, A., Hoover, E. A., Grove, D. M., Holmes, E. C., & Parrish, C. R. (2014). Host-specific parvovirus evolution in nature is recapitulated by in vitro adaptation to different carnivore species. PLoS Pathogens, 10, 11. https://doi.org/10.1371/journal.ppat.1004475.
- Behdenna, A., Lembo, T., Calatayud, O., Cleaveland, S., Halliday, J. E. B., Packer, C., … Viana, M. (2019). Transmission ecology of canine parvovirus in a multi-host, multi-pathogen system. Proceedings of the Royal Society B: Biological Sciences, 286, 20182772. https://doi.org/10.1098/rspb.2018.2772
- Bucafusco, D., Argibay, H., Diaz, L., Vega, C., Minatel, L., Postma, G. C., … & Bratanich, A. (2019). First characterization of a canine parvovirus causing fatal disease in coatis (Nasua nasua). Archives of virology, 164(12), 3073- 3079. https://doi.org/10.1007/s00705-019-04417-4
- Buonavoglia, C., Martella, V., Pratelli, A., Tempesta, M., Cavalli, A., Buonavoglia, D., … & Carmichael, L. (2001). Evidence for evolution of canine parvovirus type 2 in Italy. Journal of General Virology, 82(12), 3021-3025. https://doi.org/10.1099/0022-1317-82-12-3021
- Castillo, C., Neira, V., Aniñir, P., Grecco, S., R Pérez., Panzera, … & Ortega, R. (2020). First molecular identification of canine parvovirus type 2 (CPV2) in Chile reveals high occurrence of CPV2c antigenic variant. Frontiers in Veterinary Science, https://doi.org/10.3389/fvets.2020.00194
- Chen, C. C., Chang, A. M., Wada, T., Chen, M. T., Tu, Y. S. (2019). Distribution of Carnivore protoparvovirus 1 in free-living leopard cats (Prionailurus bengalensis chinensis) and its association with domestic carnivores in Taiwan. PLoS One, 14, e0221990. https://doi.org/10.1371/journal.pone.0221990
- Cotmore, S. F., Agbandje-McKenna, M., Canuti, M., Chiorini, J. A., Eis-Hubinger, A. M., Hughes, J., … Ictv Report Consortium. (2019). ICTV Virus Taxonomy Profile: Parvoviridae. Journal of General Virology, 100, 367-368. https://doi.org/10.1099/jgv.0.001212
- Cotmore, S. F., Agbandje-McKenna, M., Chiorini, J. A., Mukha, D. V., Pintel, D. J., Qiu, J., … Davison, A. J. (2014). The family Parvoviridae. Archives of Virology, 159, 1239-1247. https://dx.doi.org/10.1007%2Fs00705-013-1914-1
- De Almeida Curi, N. H., Coelho, C. M., de Campos Cordeiro Malta, M., Magni, E. M., Sábato, M. A., Araújo, A. S., … & de Souza, S.L. (2012). Pathogens of wild maned wolves (Chrysocyon brachyurus) in Brazil. Journal of Wildlife Disease, 48(4), 1052- 1056. https://doi.org/10.7589/2011-10-304.
- Decaro, N., & Buonavoglia, C. (2012). Canine parvovirus—a review of epidemiological and diagnostic aspects, with emphasis on type 2c. Veterinary Microbiology, 155(1), 1- 12. https://doi.org/10.1016/j.vetmic.2011.09.007.
- Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research, 32(5), 1792-1797. https://doi.org/10.1093/nar/gkh340
- Gallo-Calderón, M., Romanutti, C., Wilda, M., D’Antuono, A., Keller, L., Giacomodonato, M. N., … & La Torre, J. (2015). Resurgence of canine parvovirus 2a strain in the domestic dog population from Argentina. Journal of Virological Methods, 222, 145-149. https://doi.org/10.1016/j.jviromet.2015.06.012
- Garigliany, M., Gilliaux, G., Jolly, S., Casanova, T., Bayrou, C., Gommeren, K., … & Peeters, D. (2016). Feline panleukopenia virus in cerebral neurons of young and adult cats. BMC Veterinary Research, 12(1), 28. https://doi.org/10.1186/s12917-016-0657-0.
- Grecco, S., Iraola, G., Decaro, N., Alfieri, A., Alfieri, A., Gallo Calderón, M., … & Marandino, A. (2018). Inter-and intracontinental migrations and local differentiation have shaped the contemporary epidemiological landscape of canine parvovirus in South America. Virus evolution, 4(1), vey011. https://doi.org/10.1093/ve/vey011
- Ikeda, Y., Mochizuki, M., Naito, R., Nakamura, K., Miyazawa, T., Mikami, T., & Takahashi, E. (2000). Predominance of canine parvovirus (CPV) in unvaccinated cat populations and emergence of new antigenic types of CPVs in cats. Virology, 278(1), 13-19. https://doi.org/10.1006/viro.2000.0653
- Larsson, A. (2014). AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics, 30(22), 3276-3278. https://doi.org/10.1093/bioinformatics/btu531
- Martino, P. E., Montenegro, J. L., Preziosi, J. A., Venturini, C., Bacigalupe, D., Stanchi, N. O., & Bautista, E.L. (2004). Serological survey of selected pathogens of free-ranging foxes in southern Argentina, 1998-2001. Revue Scientifique et Tecnique, 23(3), 801-806. https://doi.org/10.20506/rst.23.3.1521
- Miller, M. A., Pfeiffer, W., & Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In 2010 gateway computing environments workshop (GCE), 1-8. https://doi.org/10.1109/GCE.2010.5676129
- Mora, M., Napolitano, C., Ortega, R., Poulin, E., & Pizarro-Lucero, J. (2015). Feline immunodeficiency virus and feline leukemia virus infection in free-ranging guignas (Leopardus guigna) and sympatric domestic cats in human perturbed landscapes on Chiloé Island, Chile. Journal of wildlife diseases, 51(1), 199-208. https://doi.org/10.7589/2014-04-114
- Nakamura, K., Sakamoto, M., Ikeda, Y., Sato, E., Kawakami, K., Miyazawa, T., … & Mochizuki, M. (2001). Pathogenic potential of canine parvovirus types 2a and 2c in domestic cats. Clinical and Diagnostic Laboratory Immunology, 8(3), 663-668. https://doi.org/10.1128/CDLI.8.3.663-668.2001
- Napolitano, C., Gálvez, N., Bennett, M., Acosta-Jamett, G. & Sanderson, J. 2015. Leopardus guigna. The IUCN Red List of Threatened Species 2015: e.T15311A50657245. https://dx.doi.org/10.2305/IUCN.UK.2015-2.RLTS.T15311A50657245.en. Downloaded on 01 May 2020.
- Napolitano, C., Johnson, W. E., Sanderson, J., O’Brien, S. J., Hoelzel, A. R., Freer, R., … & Poulin, E. (2014). Phylogeography and conservation genetics of the smallest felid in the American continent, the guigna (Leopardus guigna). Conservation Genetics, 15, 631- 653. https://doi.org/10.1007/s10592-014-0566-3
- Oosthuizen, A., Brettschneider, H., Dalton, D. L., Du Plessis, E. C., Jansen, R., Kotze, A., & Mitchell, E. P. (2019). Canine parvovirus detected from a serval (Leptailurus serval) in South Africa. Journal of the South African Veterinary Association, 90(1), 1-6. https://doi.org/10.4102/jsava.v90i0.1671
- Pérez, R., Calleros, L., Marandino, A., Sarute, N., Iraola, G., Grecco, S., … & Carrau, L. (2014). Phylogenetic and genome-wide deep-sequencing analyses of canine parvovirus reveal co-infection with field variants and emergence of a recent recombinant strain. PLoS One, 9(11). https://doi.org/10.1371/journal.pone.0111779
- Posada, D. (2008). jModelTest: Phylogenetic Model Averaging. Molecular Biology and Evolution. 25(7), 1253–1256. https://doi.org/10.1093/molbev/msn083
- Reed, A. P., Jones, E. V., & Miller, T. J. (1988). Nucleotide sequence and genome organization of canine parvovirus. Journal of virology, 62(1), 266-276.
- Shackelton, L. A., Parrish, C. R., Truyen, U., & Holmes, E. C. (2005). High rate of viral evolution associated with the emergence of carnivore parvovirus. Proceedings of the National Academy of Sciences, 102(2), 379-384. https://doi.org/10.1073/pnas.0406765102
- Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30(9), https://doi.org/1312-1313. 10.1093/bioinformatics/btu033
- Steinel, A., Munson, L., van Vuuren, M., & Truyen, U. (2000). Genetic characterization of feline parvovirus sequences from various carnivores. Journal of General Virology, 81(2), 345-350. https://doi.org/10.1099/0022-1317-81-2-345
- Truyen, U., & Parrish, C. R. (1992). Canine and feline host ranges of canine parvovirus and feline panleukopenia virus: distinct host cell tropisms of each virus in vitro and in vivo. Journal of virology, 66(9), 5399-5408.
- Truyen, U., Evermann, J. F., Vieler, E., & Parrish, C. R. (1996). Evolution of canine parvovirus involved loss and gain of feline host range. Virology, 215(2), 186-189.
- Uhart, M. M., Rago, M. V., Marull, C. A., Ferreyra H. del V., Pereira J. A. (2012). Exposure to selected pathogens in to selected pathogens in Geoffroy’s cats and domestic carnivores from central Argentina. Journal of Wildlife Disease, 48(4), 899- 909. https://doi.org/10.7589/2011-05-137.
- Viscardi, M., Santoro, M., Clausi, M. T., Cozzolino, L., Decaro, N., Colaianni, M. L., & Fusco, G. (2019). Molecular detection and characterization of carnivore parvoviruses in free‐ranging Eurasian otters (Lutra lutra) in southern Italy. Transboundary and emerging diseases, 66(5), 1864-1872. https://doi.org/10.1111/tbed.13212
- Wang S. L., Tu Y. C., Lee M. S., Wu L. H., Chen T. Y., Wu C. H., …, & Li W. T. (2019). Fatal canine parvovirus-2 (CPV-2) infection in a rescued free-ranging Taiwanese pangolin (Manis pentadactyla pentadactyla). Transboundary and Emerging Disease. 2019 Dec 30. https://doi.org/10.1111/tbed.13469
- Wang, K., Du, S., Wang, Y., Wang, S., Luo, X., Zhang, Y., … & Hu, G. (2019). Isolation and identification of tiger parvovirus in captive siberian tigers and phylogenetic analysis of VP2 gene. Infection, Genetics and Evolution, 75, 103957. https://doi.org/10.1016/j.meegid.2019.103957
Figure and Table Legends
Table 1. Sample identification used in this study. Identification of accession number, strain name, date collection, city, virus type were included.
Table 2. Amino acid variations in VP2 in samples analysed in this study.
Table 3. Amino acid variations in NS1 in samples analysed in this study.
Figure 1 . Phylogenetic analysis of the CPV complete coding region. The database was constructed using the sequences obtained in this study and all CPV sequences available in GenBank. The phylogenetic tree was inferred using the maximum likelihood method in RaxML based on GTR+I+G4 nucleotide substitution model with 1000 bootstrap replicates. Bootstrap values are indicated in the main clades. Chilean domestic dog, guiña, reference domestic dog, and other species CPV strains are represented in different colors.

No hay comentarios:
Publicar un comentario